AWS HPC Blog
Data Science workflows at insitro: how redun uses the advanced service features from AWS Batch and AWS Glue
This post was contributed by Matt Rasmussen, VP of Software Engineering at insitro.
In my first post on our data science tool, we introduced redun‘s basic features for a common Bioinformatic workflow, sequence read alignment, using an example redun workflow (06_bioinfo_batch).
Here, we will continue to use this example to describe how redun makes use of specific AWS features, such as AWS Batch’s Array Jobs and AWS Glue’s DynamicFrame, to scale large computationally heterogenous workflows.
Building bigger workflows, hierarchically
In the previous blog post of this series, we showed how to combine redun tasks to create a pipeline, align_sample, for aligning the DNA sequencing reads of a single patient sample to the reference Human genome. Now, we can continue to define even higher-level tasks. For example, in the task align_samples, we perform read alignment for a whole collection of patient samples.
@task()
def align_samples(
sample_read_pairs: Dict[str, Dict[str, str]],
genome_ref_file: File,
genome_ref_index: Dict[str, File],
sites_files: List[File],
output_path: str,
) -> Dict[str, Dict[str, Any]]:
"""
Perform alignment for sample read pairs.
"""
return {
sample_id: align_sample(
sample_id,
read_pairs,
genome_ref_file,
genome_ref_index,
sites_files,
os.path.join(output_path, sample_id),
)
for sample_id, read_pairs in sample_read_pairs.items()
}
Even though this appears to be a simple task, there are several very useful features being used here.
First, we are calling align_sample within a dictionary comprehension. The redun scheduler recurses into the five main Python container types (list, dict, set, tuple, NamedTuple) looking for Expressions to evaluate. In this case, redun will find multiple Expressions for executing align_sample, which can range from dozens to thousands of samples we would like to process. redun’s scheduler will automatically detect that these Expressions are independent of one another and thus are safe to execute in parallel (Figure 1, center). This allows users to define parallel execution using a very natural Python syntax. The expressions for align_sample will in turn be evaluated to ultimately obtain the aligned sequences (BAM files) as we saw previously.
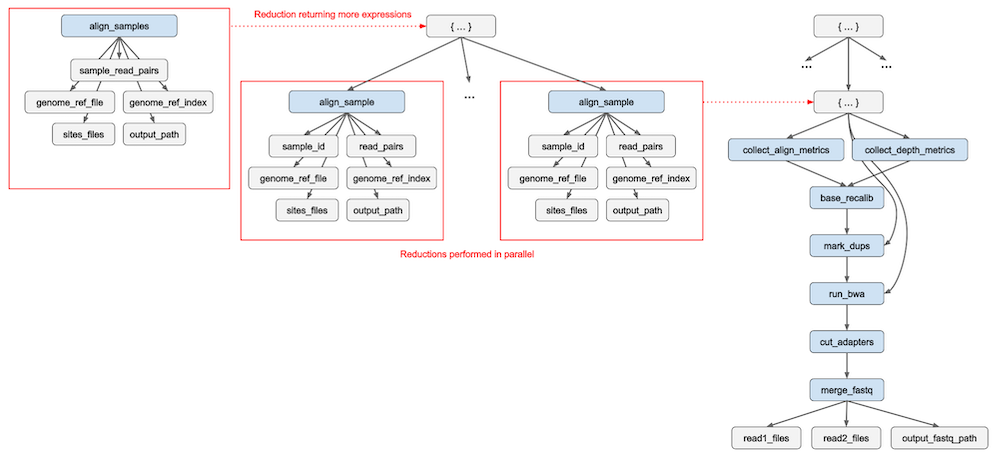
Figure 1: Graph reduction for evaluating subworkflows. When the Expression align_samples is reduced (left) it returns a Python dictionary containing additional Expressions (align_sample). The redun scheduler will recurse into task return values to find more potential reductions. If multiple subtrees are ready for reduction (center), their executions can be done automatically in parallel. The Expression Graph can expand and contract until a single concrete value is obtained.
Optimizations: AWS Array Jobs
Sometimes workflows will have very large fan-outs, such as processing tens of thousands of samples, and if implemented naively it would lead to thousands of AWS Batch and S3 API calls just to submit the work, leading to a lack of parallelism. Here, redun’s implementation makes use of a convenient feature of AWS Batch called Array Jobs.
To review, AWS Array Jobs allows using a single AWS Batch submit API call to submit up to 10,000 Batch jobs at once. However, this feature comes with a few conditions. First, all of the jobs must share a common Batch Job Definition, including a common top-level command. Second, the jobs will each have an environment variable defined AWS_BATCH_JOB_ARRAY_INDEX that gives the job its index in the array. Using this index, the job must then customize its execution.
When a large fan-out is encountered in a redun workflow, redun will automatically make use of Array Jobs transparently to the user in order to efficiently submit work to the AWS Batch cluster. It achieves this using the following technique.
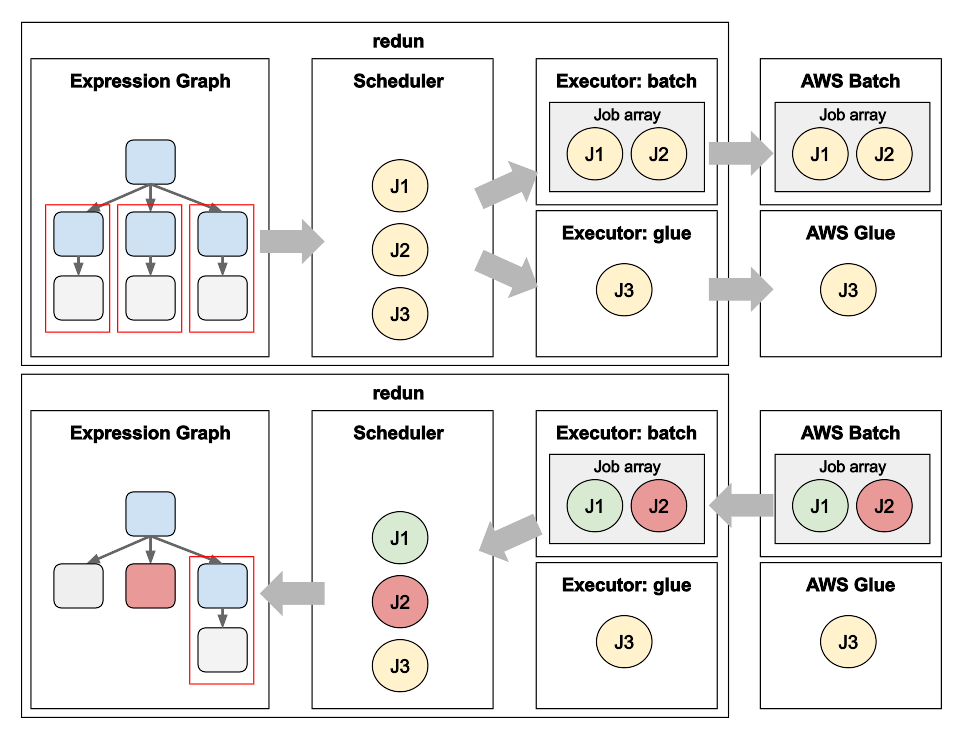
Figure 2: Identification of redun jobs and grouping them into Array Jobs to run on AWS Batch. (Top) redun represents the workflow as an Expression Graph (top-left), and identifies reductions (red boxes) that are ready to be executed. The redun Scheduler creates a redun Job (J1, J2, J3) for each reduction and dispatches those jobs to Executors based on the task-specific configuration. The Batch Executor allows jobs to accumulate for up to three seconds (default) in order to identify compatible jobs for grouping into an Array Job, which are then submitted to AWS Batch (top-right). (Bottom) As jobs complete in AWS Batch, the success (green) and failure (red) is propagated back to Executors, the Scheduler, and eventually substituted back into the Expression Graph (bottom-left).
First, when the redun Scheduler encounters an Expression that needs evaluation, it creates a redun Job, inspects which executor the Expression is requesting, and submits the redun Job to the executor (Figure 2). For AWS Batch, redun has implemented a AWSBatchExecutor which receives redun Job submissions and in turn creates and submits AWS Batch Jobs. However, instead of submitting each job immediately to AWS Batch, we employ a small delay (typically three seconds by default), and store jobs for submission into an in-memory queue. After the delay expires, we then inspect all jobs within the queue and automatically group them into Array Jobs. Specifically, we look for jobs that make use of the same Job Definition, which is especially common for large fan-outs.
Redun also prepares a specialized S3 input file for each array. Typically, task arguments are serialized for a redun Job in the S3 scratch space (defined by s3_scratch in redun.ini). However, if that was done for every job, it would lead to thousands of S3 API calls and a slowdown in overall job submission. Instead, redun bundles all input arguments for the entire Array Job into a single serialized list stored in S3. When each Batch Job runs, it will use its AWS_BATCH_JOB_ARRAY_INDEX environment variable to index into the input array to find its appropriate arguments.
Overall, these techniques allow us to scale down the number of API calls involved in a fan out by a factor up to 10,000. Redun is also able to automatically make use of these features without requiring users to use any specialize syntax in their workflows.
Mixed compute infrastructure
In the Bioinformatics example above, we discussed how redun can submit jobs to AWS Batch. However, in general each task can run in its own thread, process, Docker container, AWS Batch job, or AWS Glue job. Users can lightly annotate where they would like each task to run (e.g. executor="batch" or executor="glue"), and redun will automatically handle the data and code movement as well as backend scheduling.
For example, we may want a portion of our workflow to perform Spark calculations. In the example code below, we show a common Cheminformatics task of taking molecules described in the SMILES format and deriving their INCHI identifiers. To do this efficiently for large virtual libraries of molecules (billions of molecules), we configure our redun task to use the “glue” Executor, which instructs the redun scheduler to package our Python code to run on AWS Glue. When running in AWS Glue, our task will have access to the SparkContext, which allows performing typical Spark calculations. In this example, we load a sharded dataset on S3 with molecules into a RDD (resilient distributed dataset), and compute in parallel using a UDF (user defined function) the INCHI identifiers.
from rdkit import Chem
from redun import glue, task, ShardedS3Dataset
@glue.udf
def get_inchi(smiles: str) -> str:
"""
Spark UDF (user defined function) that can be used to map the rows of an RDD.
"""
return Chem.inchi.MolToInchi(Chem.MolFromSmiles(smiles))
@task(executor="glue", additional_libs=["rdkit-pypi"], workers=5, worker_type="G.1X")
def calculate_inchi(
input_ds: ShardedS3Dataset, output_dir: str, smiles_col: str = "smiles"
) -> ShardedS3Dataset:
"""
Adds the "inchi" column to a DataFrame as calculated from a column containing SMILES strings.
Example:
input_ds = SharedS3Dataset("s3://YOUR_BUCKET/library_with_smiles")
output_ds = calculate_inchi(input_ds, "s3://YOUR_BUCKET/library_with_inchi", "smiles")
"""
# Load sharded dataset into a Spark RDD.
rdd = input_dir.load_spark()
# Generate INCHI column efficiently using a Spark User Defined Function (UDF).
rdd_inchi = dataset.withColumn("inchi", get_inchi(smiles_col))
# Save dataset.
output_ds = ShardedS3Dataset.from_data(rdd_inchi, output_dir)
return output_ds
Note, the task calculate_inchi can be called from any other redun task, regardless of the executor. So we are free to mix Glue with local or other Batch compute into a comprehensive end-to-end workflow.
Conclusion
In this blog series, we reviewed a new data science tool, redun, that allows easy development of scientific workflows in the cloud using serverless compute platforms like AWS Batch and AWS Glue. Specifically, we showed how redun lends itself to Bioinformatics workflows which typically involve wrapping Unix-based programs that require file staging to and from object storage. We also showed how redun leverages AWS Batch-specific features like Array Jobs to support workflows with large fan-outs. However, if some parts of your compute requires Spark, we showed how redun can mix compute backends together into a single workflow. We hope by open sourcing redun, we can help other data science teams easily develop complex workflows in the cloud and do great science.
The content and opinions in this blog are those of the third-party author and AWS is not responsible for the content or accuracy of this blog.