AWS HPC Blog
Category: Compute
Introducing the Spack Rolling Binary Cache hosted on AWS
Today we’re excited to announce the availability of a new public Spack Binary Cache. In a collaboration, between AWS, E4S, Kitware, and the Lawrence Livermore National Laboratory (LLNL), Spack users now have access to a public build cache hosted on Amazon S3. The use of this Binary Cache will result in up to 20x faster install times for common Spack packages.
Encoding workflow dependencies in AWS Batch
This post covers the different ways you can encode a dependency between basic and array jobs in AWS Batch. We also cover why you may want to encode dependencies outside of Batch altogether using a workflow system like AWS Step Functions or Apache Airflow.
AWS Batch updates: higher compute utilization, AWS PrivateLink support, and updatable compute environments
In this post, I cover some of the recent updates to AWS Batch, including improvements to job placement, addition of AWS PrivateLink support, and the new capabilities to update your AWS Batch compute environments.
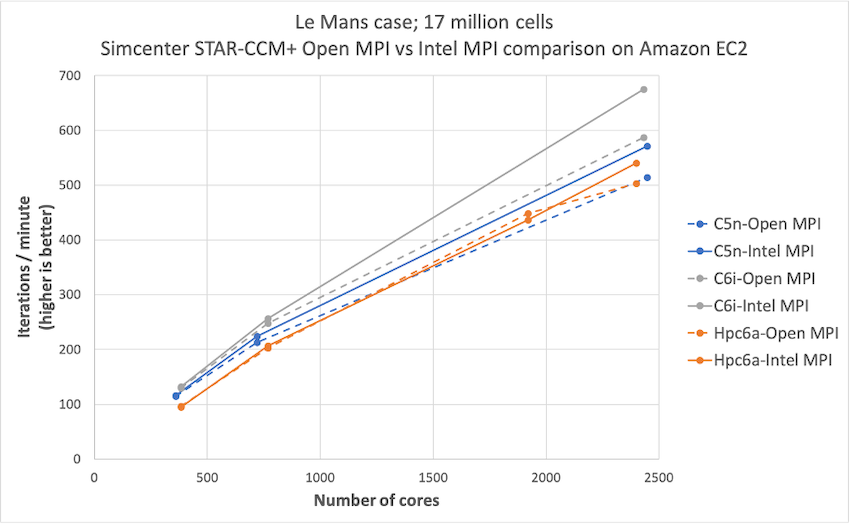
Simcenter STAR-CCM+ price-performance on AWS
Organizations such as Amazon Prime Air and Joby Aviation use Simcenter STAR-CCM+ for running CFD simulations on AWS so they can reduce product manufacturing cycles and achieve faster times to market. In this post today, we describe the performance and price analysis of running Computational Fluid Dynamics (CFD) simulations using Siemens SimcenterTM STAR-CCM+TM software on AWS HPC clusters.
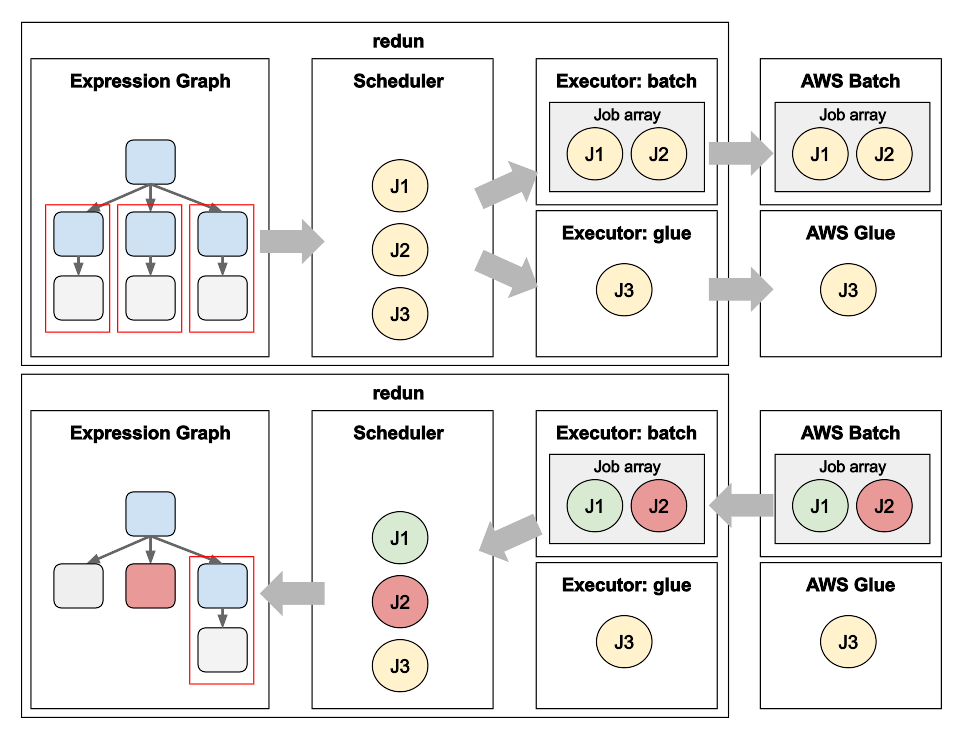
Data Science workflows at insitro: how redun uses the advanced service features from AWS Batch and AWS Glue
Matt Rasmussen, VP of Software Engineering at insitro, expands on his first post on redun, insitro’s data science tool for bioinformatics, to describe how redun makes use of advanced AWS features. Specifically, Matt describes how AWS Batch’s Array Jobs is used to support workflows with large fan-out, and how AWS Glue’s DynamicFrame is used to run computationally heterogenous workflows with different back-end needs such as Spark, all in the same workflow definition.
Data Science workflows at insitro: using redun on AWS Batch
Matt Rasmussen, VP of Software Engineering at insitro describes their recently released, open-source data science framework, redun, which allows data scientists to define complex scientific workflows that scale from their laptop to large-scale distributed runs on serverless platforms like AWS Batch and AWS Glue. I this post, Matt shows how redun lends itself to Bioinformatics workflows which typically involve wrapping Unix-based programs that require file staging to and from object storage. In the next blog post, Matt describes how redun scales to large and heterogenous workflows by leveraging AWS Batch features such as Array Jobs and AWS Glue features such as Glue DynamicFrame.
Migrating to AWS ParallelCluster v3 – Updated CLI interactions
The AWS ParallelCluster version 3 CLI differs significantly from ParallelCluster version 2. This post provides some guidance on mapping between versions to help you with migrating to ParallelCluster 3. We also summarize new CLI features in ParallelCluster 3 to expose the things you just couldn’t do previously.
Choosing between AWS Batch or AWS ParallelCluster for your HPC Workloads
It’s an understatement that AWS has a lot of services (more than 200 at the time of this post!). We’re usually the first to point out that there’s more than one way to solve a problem. HPC is no different in this regard, because we offer a choice: customers can run their HPC workloads using AWS […]
Optimize your Monte Carlo simulations using AWS Batch
Introduction Monte Carlo methods are a class of methods based on the idea of sampling to study mathematical problems for which analytical solutions may be unavailable. The basic idea is to create samples through repeated simulations that can be used to derive approximations about a quantity we’re interested in, and its probability distribution. In this […]
GROMACS performance on Amazon EC2 with Intel Ice Lake processors
We recently launched two new Amazon EC2 instance families based on Intel’s Ice Lake – the C6i and M6i. These instances provide higher core counts and take advantage of generational performance improvements on Intel’s Xeon scalable processor family architectures. In this post we show how GROMACS performs on these new instance families. We use similar methodologies as for previous posts where we characterized price-performance for CPU-only and GPU instances (Part 1, Part 2, Part 3), providing instance recommendations for different workload sizes.