AWS News Blog
Vodafone DreamLab – Accelerating Cancer Research
Continuing what is quickly turning in to series of customer stories, I would like to turn the keyboard over to our friends at Vodafone!
— Jeff;
You may have seen an interesting application called DreamLab circulating on the internet and social media in the past couple of weeks, and we thought we’d give you some insight into how this very innovative application works under the hood.
Fighting Cancer
Cancer touches so many of us; with one in two Australians being diagnosed with cancer by their 85th birthday, medical research is the key to finding better treatments.
Cancer research progress at Garvan Institute of Medical Research is slowed by the limited access researchers have to the supercomputers they need to crunch complex research data.
For a long time, cancer has been thought of by its tissue of origin – for example lung, breast or pancreatic cancer. But cancer is a disease of the DNA, and so with the advances in genome sequencing, world leading researchers at Garvan Institute of Medical Research are interested in creating a library of cancers, grouped based on their genetic mutations.
Analysing Mutations
To do this, researchers need to analyse the genomic mistakes (DNA mutations) of thousands of different cancer patients and group them based on their genetic profiles (not the tissue in which the cancer originated). Garvan has sourced somatic mutations from cancer patients de-identified for research purposes from the published studies of the International Cancer Genome Consortium (ICGC) and The Cancer Genome Atlas (TCGA) to do this. Whilst the research project aims to analyse 24 cancer types / subtypes, the DreamLab app enables users to choose to support breast, pancreatic, ovarian or prostate cancer as the first stage of the project.
The sequencing of just one genome generates tens of gigabytes of data. So huge computing power is needed to complete this analysis. But at Garvan (and around the world) access to supercomputers is limited and costly. That’s where Vodafone’s DreamLab comes in – it enables everyone to donate the computer processing power of their idle Smartphones to speed up cancer research. At the moment the app is available on Android only.
To tackle this problem the Garvan Institute of Medical Research has developed a novel in-house computational algorithm to estimate how functionally close the mutated genes between two patients could be, which can then be used to group tumours based on their mutation profiles, regardless of which tissue the cancer originated.
This algorithm, called the Network Connectivity Analyser (NCA) – was developed by Dr Hong Ching Lee and Dr Jianmin Wu from The Garvan Institute of Medical Research. It has been adapted to run on Android Smartphones for DreamLab.
Inside DreamLab
 The algorithm calculates a number of statistics of the interactions between two sets of genes (i.e. mutated genes from two patients). Within the DreamLab app, the NCA will perform the cross comparison processing whenever the phone is charged to at least 95%. These patterns can help researchers identify subgroups of patients who share similar mutation profiles, and could therefore potentially respond to the same therapies. The combination of a large community (DreamLab users), coupled with big data analytic algorithms, makes this revolutionary form of research possible.
The algorithm calculates a number of statistics of the interactions between two sets of genes (i.e. mutated genes from two patients). Within the DreamLab app, the NCA will perform the cross comparison processing whenever the phone is charged to at least 95%. These patterns can help researchers identify subgroups of patients who share similar mutation profiles, and could therefore potentially respond to the same therapies. The combination of a large community (DreamLab users), coupled with big data analytic algorithms, makes this revolutionary form of research possible.
Here’s how DreamLab works:
- Garvan uploads their large research problem to Amazon Simple Storage Service (Amazon S3).
- Once a user has downloaded and set up the DreamLab app, the app will then authenticate against Amazon Cognito and use a set of temporary, limited privilege credentials to request a research job out of Amazon SQS, and then download a small research payload out of S3 (hundreds of KB) with job session state for each phone being managed by Amazon DynamoDB.
- A novel algorithm (built by Garvan researchers) in the DreamLab app, allows the phone to solve the research problem, using the phone’s computing power (the algorithm enables the comparison of the functional similarities and differences in mutated genes from different patients, to enable creation of this library of cancers grouped by their genetic profile).
- The result is then sent back from the phone to S3, for the Garvan team to analyse.
Informally, this is similar to solving a crossword puzzle with everyone working on a different clue.
Architecture Rationale
Here’s how DreamLab uses AWS:
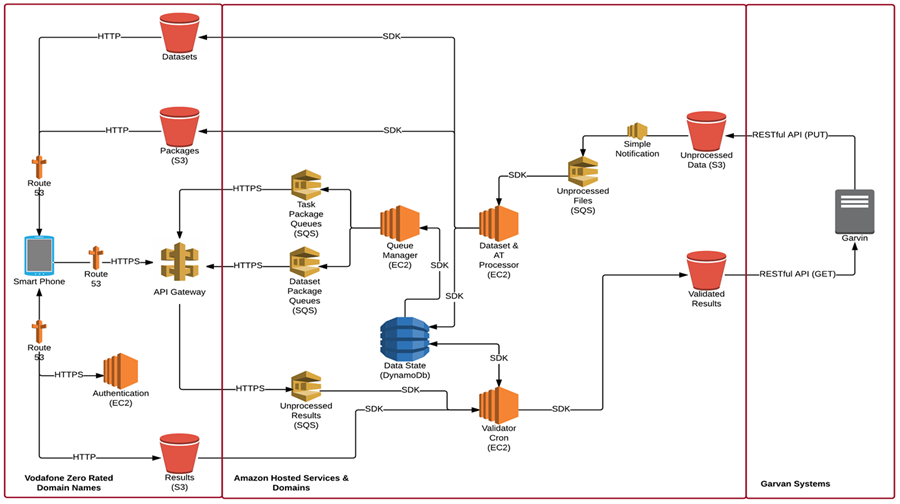
The nature of the project required an architecture that supports large volumes of data and spiked traffic, whilst remaining cost effective. This required services that can auto scale and/or have no capacity limits. The architecture also needed to support the ability to maintain the state of any data item, whilst simultaneously being updated across more than one client.
Amazon Simple Storage Service (Amazon S3) is an optimal service for the storage of data. It has no upper limit in the amount of data that can be stored, and due to its distributed nature it has high redundancy. It also has the added functionality of being able to fire events when an item is added.
Amazon Simple Queue Service (Amazon SQS) is a queuing system of unlimited size, supporting the requirement of high traffic and data. The app facing queues utilise the data visibility setting, to ensure that other devices do not fetch an item whilst various operations are taking place.
Amazon DynamoDB is a highly scalable NoSQL database that has no upper limit in record quantity. Whilst the data schema for this system could benefit from a relational database syste,m the benefits of the scale and price of DynamoDB outweigh these.
Amazon Cognito and STS provide substantial security with only a minimal requirement for custom development.
Amazon Elastic Compute Cloud (Amazon EC2) servers run all the custom code that is used to maintain the other systems. This custom code is scheduled to run regularly with CRON jobs. The EC2 servers are limited in capacity, but as they are working behind SQS this is not be an issue. We investigated AWS Lambda, a service that runs custom code on an event notification, however Lambda is currently not available in Australia.
The app accesses all SQS queues through Amazon API Gateway. This is so that the communication is over a custom domain that Vodafone uses to zero rate data charges for Vodafone Australia customers in Australia.
By the Numbers
The Garvan Institute currently has 100,000 base datasets files. Each file is 2 MB uncompressed, and 500 KB compressed. In the DreamLab project, each of the base datasets will be downloaded by three users for result validation purposes.
Garvan also has 5,000 analysis tasks, with a size of 1 KB uncompressed and 250 bytes compressed.
Based on the team’s tests, 33 new Android devices will analyse the same amount data about as fast as a CPU core of the Garvan’s supercomputer. The Garvan’s supercomputer consists of 1,280 CPU cores in total.
As of today, DreamLab has over 44,000 active users, which is providing over 1,000 times the processing power when compared to the Garvan’s existing supercomputer allocation for cancer research.
Congratulations & Acknowledgements
Congratulations to Vodafone Foundation, The Garvan Institute of Medical Research and the mobile application partner b2cloud on this great innovation.
You can download DreamLab from the Google Play Store and contribute to this effort if you’d like!
— Andrew Burnet, Domain Delivery Lead, Vodafone Australia