AWS for Industries
How Vertex Pharmaceuticals reduced time and cost of small molecule drug discovery using Thompson Sampling on AWS
This post was co-written with Roberto Iturralde, Sr. Director of Engineering, and Rajarshi Guha, Director of Data and Computational Sciences at Vertex Pharmaceuticals.
Vertex is a global biotechnology company that invests in scientific innovation to create transformative medicines for people with serious diseases. In this blog post, we’ll show how Vertex Pharmaceuticals designed a highly performant, and cost optimized small molecule search system on AWS.
Overview
The modern drug discovery process is increasingly a data-driven effort, where multiple steps in the discovery pipeline involve instruments, systems, or devices that generate large volumes of data that scientists use to answer complex questions about the biology and chemistry underlying a disease. Datacenter environments can be challenged to meet these needs, where scientific teams need short term access to hundreds of terabytes of storage or thousands of CPUs. Speed and efficiency are critical at each stage of the drug discovery process both because the resources involved are costly and, most importantly, patients are waiting for a therapy to improve their lives.
For disease programs where Vertex Pharmaceuticals determines that a small molecule is a suitable therapeutic approach, one of the initial steps is to identify small molecules that might address the underlying biology. Given that the possible number of pharmacologically relevant small molecules is larger than the number of stars in the observable universe, experimental testing is infeasible. Instead, Vertex employs, among other strategies, computational methods to evaluate billions of chemical structures and prioritize a subset of compounds for experimental testing. Vertex Pharmaceuticals scientists employ several methods to analyze these massive collections which range from computing shape similarity between two small molecules to testing whether a small molecule can “dock” to the binding site of a protein. A brute force search using any of these methods, for libraries containing billions of molecules, can take from days to years depending on the complexity of the search. For example, one common commercial vendor library contains roughly 17 billion virtual molecules. A 3D comparison averages one second per molecule per vCPU, leading to over 600 CPU-years to scan through the entire library.
Thompson Sampling
Thompson Sampling is a heuristic for choosing actions that was described in 1933, and has been applied in settings such as clinical trials and A/B testing. Thompson Sampling balances tradeoffs between exploration and exploitation and relies on Bayesian update of a prior distribution by probabilistically focusing the search over a given promising area of the search space. Vertex Pharmaceuticals scientists developed an approach to use Thompson Sampling to search molecule libraries of arbitrary size.
Architecture Overview
Vertex Pharmaceuticals had the following goals for their distributed Thompson Sampling Architecture:
- Performance – To accelerate their drug discovery process, they wanted to scale their compound search horizontally to return results as quickly as possible, even as the size of the small molecule libraries grow.
- Cost Optimization – Given the scale these compound searches can achieve and the frequency with which these searches are run, then wanted a cost-effective architecture.
- Democratized access – The computational chemists who designed the Thompson Sampling system wanted to allow it to be used by their colleagues without needing to understand the internals of the system or require AWS expertise
To meet the performance and cost goals, Vertex developed a serverless architecture using AWS Fargate Spot fleet for the compute-intensive, stateless search workers. To democratize access to this system, Vertex created a serverless API using Amazon API Gateway that dynamically creates a Thompson Sampling search environment based on details from the API request.
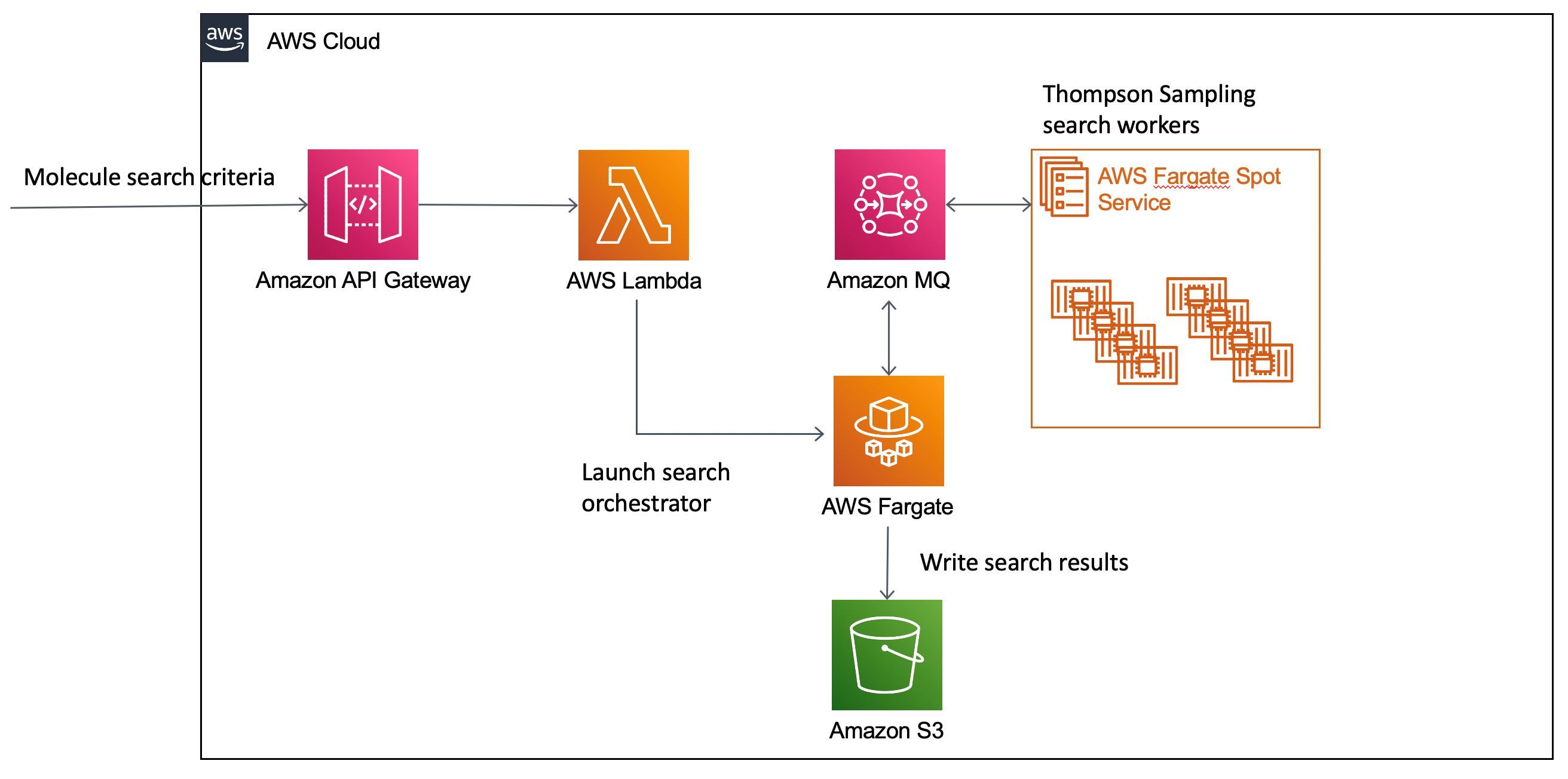
Figure 1: Small molecule search job using Thompson Sampling workflow on AWS.
- Computational chemists issue a HTTP POST to an Amazon API Gateway endpoint with the details of the molecule search.
- An AWS Lambda function behind this endpoint creates an AWS Fargate task to orchestrate the search, an Amazon MQ Rabbit queue to broker communication between the orchestrator (see below) and the workers, and submits the search request to the orchestrator.
- The orchestrator creates a Fargate Spot service of search workers sized based on the search request (typically 100-500 tasks) and communicates with the workers to drive the search via the queue. The virtual library is defined in terms of reactions and reagents, stored in a relational database. Each worker samples reagents within a given reaction to generate the actual product molecules that are then scored.
- Workers send score information for the product molecules they are evaluating back to the orchestrator via the queue. The orchestrator tracks the performance of each molecule the workers are evaluating and adjusts the search using the Thompson Sampling heuristic to pursue molecules similar to those that are scoring well.
- Once it has finished searching the molecule library, the orchestrator writes the result set of molecules matching the search query to Amazon S3 for the scientist to retrieve, and the Fargate and MQ resources gracefully shut-down.
Conclusion
Thompson Sampling searches routinely scale up to 1,000 vCPUs in minutes, and each Thompson Sampling search costs just a few hundred dollars to run compared to thousands of dollars for pre-provisioned underutilized resources on-premises. Importantly, the Thompson search implementation only needed to examine a small fraction (0.07%) of the full library, resulting in 18 to 20 fold speed up compared to a traditional serial approach. This architecture is completely serverless, scaling down to zero and costing nothing when idle. The serverless architecture also allows the engineers who partner with these scientists to spend more time delivering new features and less time managing infrastructure. On the scientific projects where the Thompson Sampling search has been used, Vertex Pharmaceuticals has been able to virtually evaluate chemical matter and prioritize a subset of the chemical matter for experimental testing. These novel findings give our scientists new areas to explore as they continue their pursuit of transformative therapeutics for our patients.
To learn more about how AWS and APN partner solutions are empowering life sciences research and discovery, visit thinkwithwp.com/health/biopharma/solutions/
About the authors
 Rajarshi Guha is a Director at Vertex Pharmaceuticals where he leads Data & Methods within the global Data and Computational Sciences group. With over 15 years of experience in handling, analyzing, and visualizing chemical information, he has developed novel algorithms for the analysis and visualization of chemical data types, large scale infrastructure projects (Pharos, BARD), and worked on small molecule screening programs for rare cancers and infectious diseases.
Rajarshi Guha is a Director at Vertex Pharmaceuticals where he leads Data & Methods within the global Data and Computational Sciences group. With over 15 years of experience in handling, analyzing, and visualizing chemical information, he has developed novel algorithms for the analysis and visualization of chemical data types, large scale infrastructure projects (Pharos, BARD), and worked on small molecule screening programs for rare cancers and infectious diseases.

Roberto Iturralde is a Senior Director of Cloud Computing at Vertex Pharmaceuticals. As a part of the Software Engineering group, Roberto is responsible for Vertex engineering’s cloud strategy and the architectures of systems built to accelerate scientific research. Roberto’s background is in application design, large scale cloud architecture, and serverless computing.