Containers
Using Amazon ECS with NVIDIA GPUs to accelerate drug discovery
This post was written in collaboration with Neel Patel, Drug Discovery Scientist, Nvidia.
Drug discovery is the process through which potential new medicines are identified. It involves a wide range of scientific disciplines, including biology, chemistry, and pharmacology, as well as computer science. AstraZeneca and NVIDIA collaborated on developing MegaMolBART so the computational drug discovery process happens in real-time and is interactive. The application searches, screens, and organizes large-scale drug-like compound databases.
Through an intuitive web-based UI, chemists can explore analogues among compounds while data scientists can perform routine cheminformatics tasks like clustering and dimensionality reduction against the RAPIDS libraries, accelerated by GPUs.
Transformers have shown the ability to embed the dense and continuous representation of chemical space, which adheres to the rules of chemistry and captures the underlying distribution of molecules in the database. This confers the correct inductive bias needed for one-shot learning and other problems where data is limited, or labels are not available.
For this solution we chose Amazon Elastic Container Service (Amazon ECS) for orchestration, since it makes it easy for customers to deploy, manage, and scale containerized applications.
Solution
In this blog we will focus on the new NVIDIA MegaMolBART solution. The solution will allow customers to explore MegaMolBART, a transformer model of the latent space of small molecules obtained through self-supervised learning of compounds represented by SMILES .
Downstream applications of these models include property prediction and novel drug-like compound generation. For more information about this project, see the GitHub repository and the NGC page.
The AWS Cloud Development Kit (AWS CDK) is an open-source software development framework to define your cloud application resources using familiar programming languages. For this solution we chose AWS CDK’s Python bindings for its simplicity.
Architecture
Architecture of the MegaMolBART solution is illustrated in the below figure.

We start with creating the base infrastructure for this solution, which includes the virtual private cloud (VPC) that spans across multiple Availability Zones with public and private subnets. Next, we create an Amazon ECS cluster with Amazon CloudWatch Container Insights enabled. We also add an AWS Cloud Map namespace for service discovery and an Amazon Elastic File System (Amazon EFS) for data persistence.
We implemented scaling at both the cluster instance level and the task level to cope with spiky loads.
Deploy NVIDIA MegaMolBART
We used AWS CDK Python library to create the resources described in the previous section. AWS CDK uses AWS CloudFormation under the hood to declaratively create resources in your AWS account. You can find the AWS CDK code and configuration templates for this project here.
Clone the project’s GitHub repository and deploy the stack.
We suggest using virtual environments with Python. You can create one by running:
After the init process completes and the virtualenv is created, you can use the following step to activate your virtualenv:
If you are using a Windows platform, you would activate the virtualenv like this:
Once the virtualenv is activated, you can install the required dependencies:
At this point you can now synthesize the AWS CloudFormation template for this code.
To deploy, run:
The deployment process takes around 20 – 25 minutes. Most of this time is spent downloading the ChEMBL database.
At the end of the deployment, you will see the URL for the CuChem service:
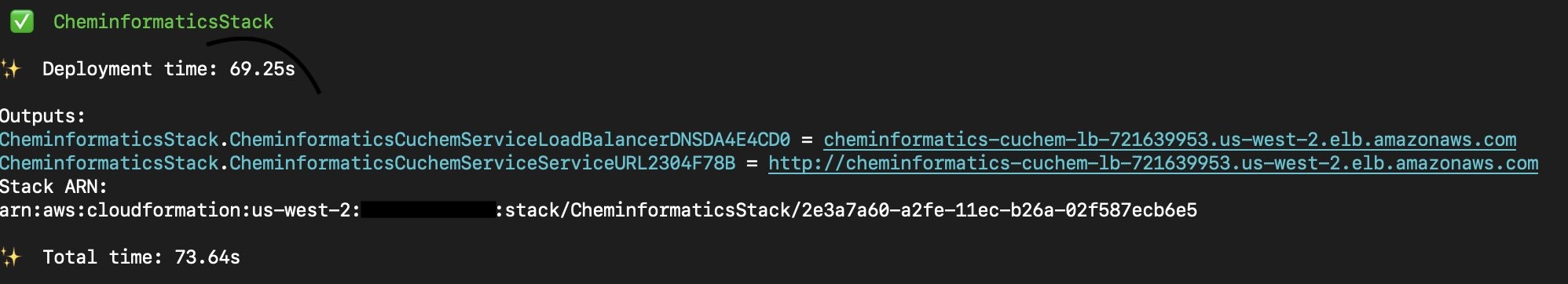
Going to that URL from your browser should take you to the application’s user interface, which would look like the following image.

For a more detailed instructions on how to use the service, check out the awesome tutorial by NVIDIA.
Monitoring
The solution uses Amazon CloudWatch Container Insights to collect, aggregate, and summarize metrics and logs. Amazon CloudWatch automatically collects metrics for containers, such as CPU, memory, disk, and network. It also provides diagnostic information, such as container restart failures, to help you isolate issues and resolve them quickly.
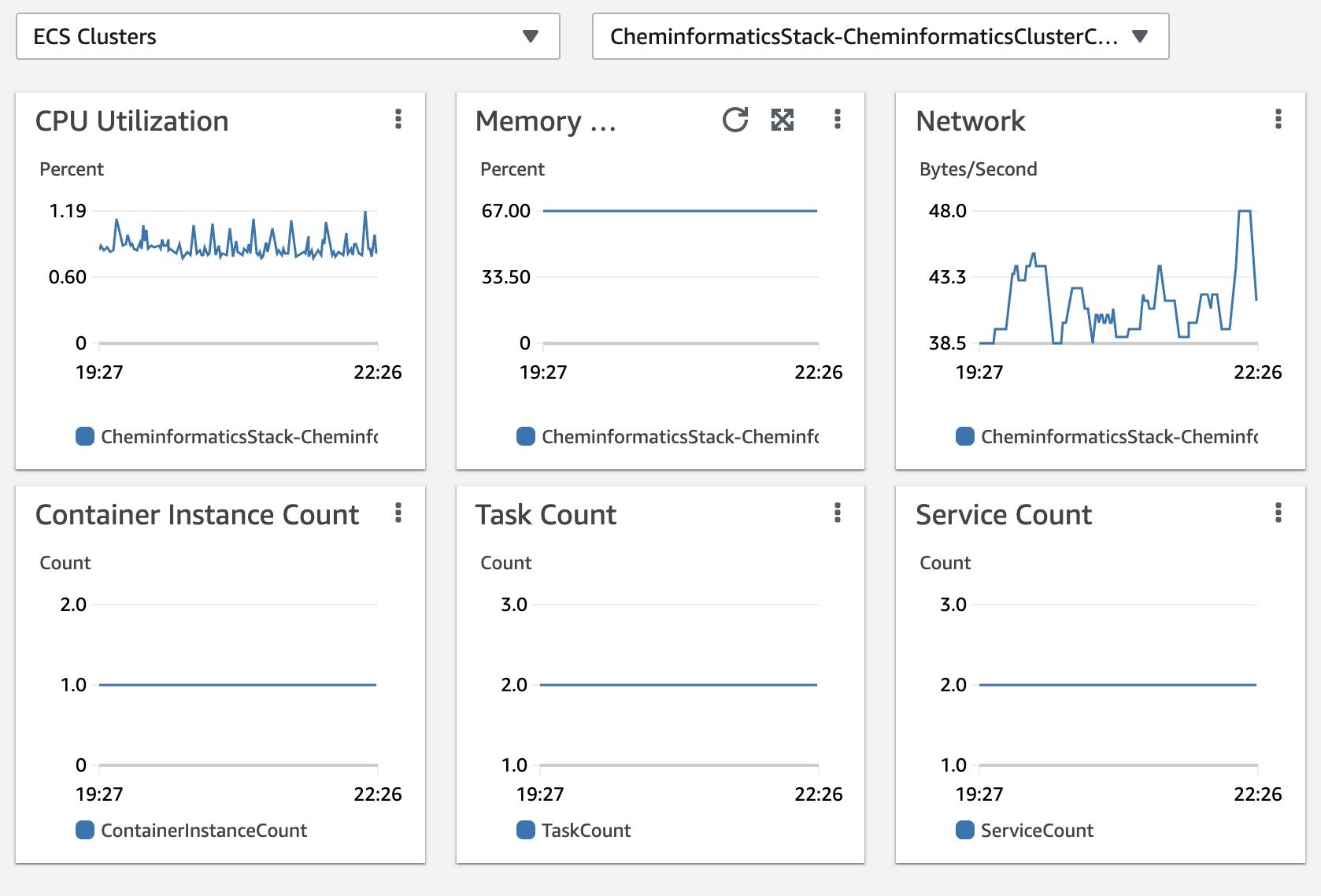
You can also set Amazon CloudWatch alarms on metrics that Container Insights collects. You can find Amazon ECS monitoring best practices here.
AWS Cloud Map for Service Discovery
Our solution includes two Amazon ECS services. The CuChem service provides the front-end for users and MegaMolBART service acts as the back-end worker service for generating novel molecules.
CuChem containers discover the address of the MegaMolBART containers using AWS Cloud Map service discovery. When the back-end workers scale, CuChem containers will automatically discover new MegaMolBART containers.
Task Placement on GPUs
Both containers that are used in this solution strictly require NVIDIA Tesla cards, which makes p3 instances a great choice. In this post, the cluster only has p3.2xlarge container instances. In case your cluster has other instance types, you can use Amazon ECS task placement constraints to ensure Amazon ECS schedules your tasks on specific Amazon Elastic Compute Cloud (Amazon EC2) instance families.
Here’s a snippet of achieving that with AWS CDK at the task level:
By adding this task placement constraint, we make sure Amazon ECS schedules the task on p3 type instances.
Bootstrapping Chembl Database
CuChem container needs the Chembl Database to operate. Whenever Amazon ECS creates a new task, it will first run the init_container container before executing other containers in the task. The init container downloads the database for CuChem and persists it on the attached Amazon EFS file system. As Amazon EFS is a shared file system, once the model data has been downloaded, subsequent spawns of this init container will skip downloading data.
The solution uses Amazon ECS ContainerDependency to define the order in which Amazon ECS should start the containers in the task.
How to Load Test
Both services, CuChem and MegaMolBART, are configured to scale automatically using Amazon ECS service auto scaling. Amazon ECS will add tasks when the aggregate CPU usage within a service exceeds 30%. We found 30% works best when it comes to reacting to spikes.
For cluster capacity autoscaling we setup step scaling since it is the recommended approach by the Amazon ECS best practices guide. We set the steps as the following: Below 15% CPU utilization 1 instance will be removed; above 30% CPU utilization 1 instance will be added; above 50% CPU utilization 1 instance will be added.
The following figure shows the relationship between the number of tasks and the number of container instances against a spiking CPU. We spin up more tasks and more instances to bear the traffic, and as the traffic scales down we scale down with it.

In order to load test our service we can use a tool such as JMeter. In our testing we found that one p3.2xlarge instance can support up to 20 simultaneous molecule generation requests. In order to handle more users, we need to add more instances using cluster autoscaling.
We created three use cases with JMeter: the first use case is simply rendering the home page; the second use case is selecting a subset of molecules; the third use case is running MegaMolBART to create novel molecules. As expected under heavy load, MegaMolBART service fails the most since it is the most intense process this application provides.
Under heavy load, we observed 5% of the MegaMolBART requests fail. Rendering the home page and selecting a subset of molecules seem to have good performance and can take heavier loads. Optimizations such as reducing the container image size and leveraging the GPU with the algorithm itself could make MegaMolBART more scalable.
Cost Analysis
AWS CDK templates included with this solution tag the resources it creates. You can use AWS Cost Explorer to monitor the usage.
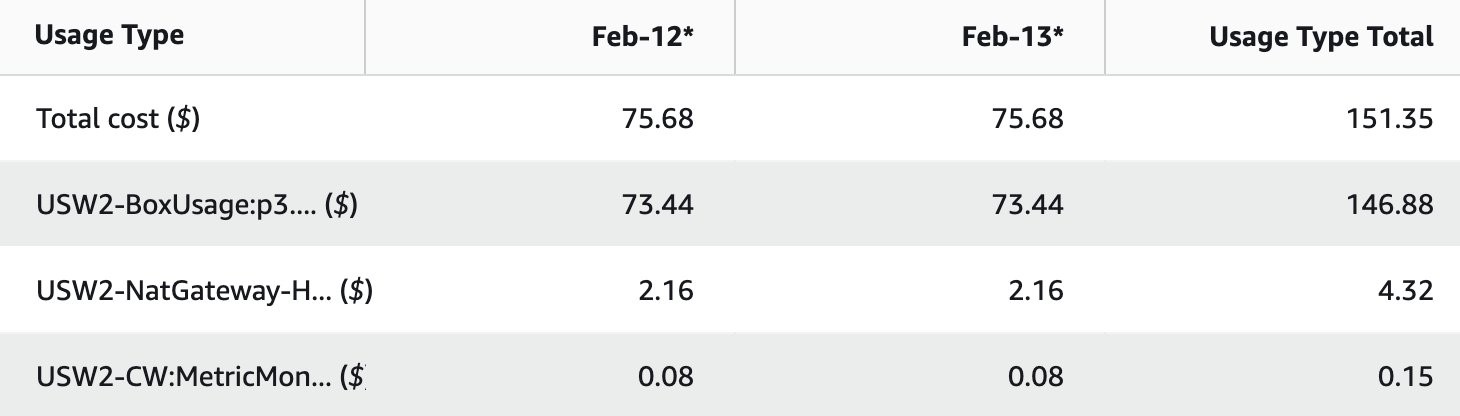
The screenshot below shows a breakdown of costs. As expected, most of the charges are for the GPU instance (p3.2xlarge).
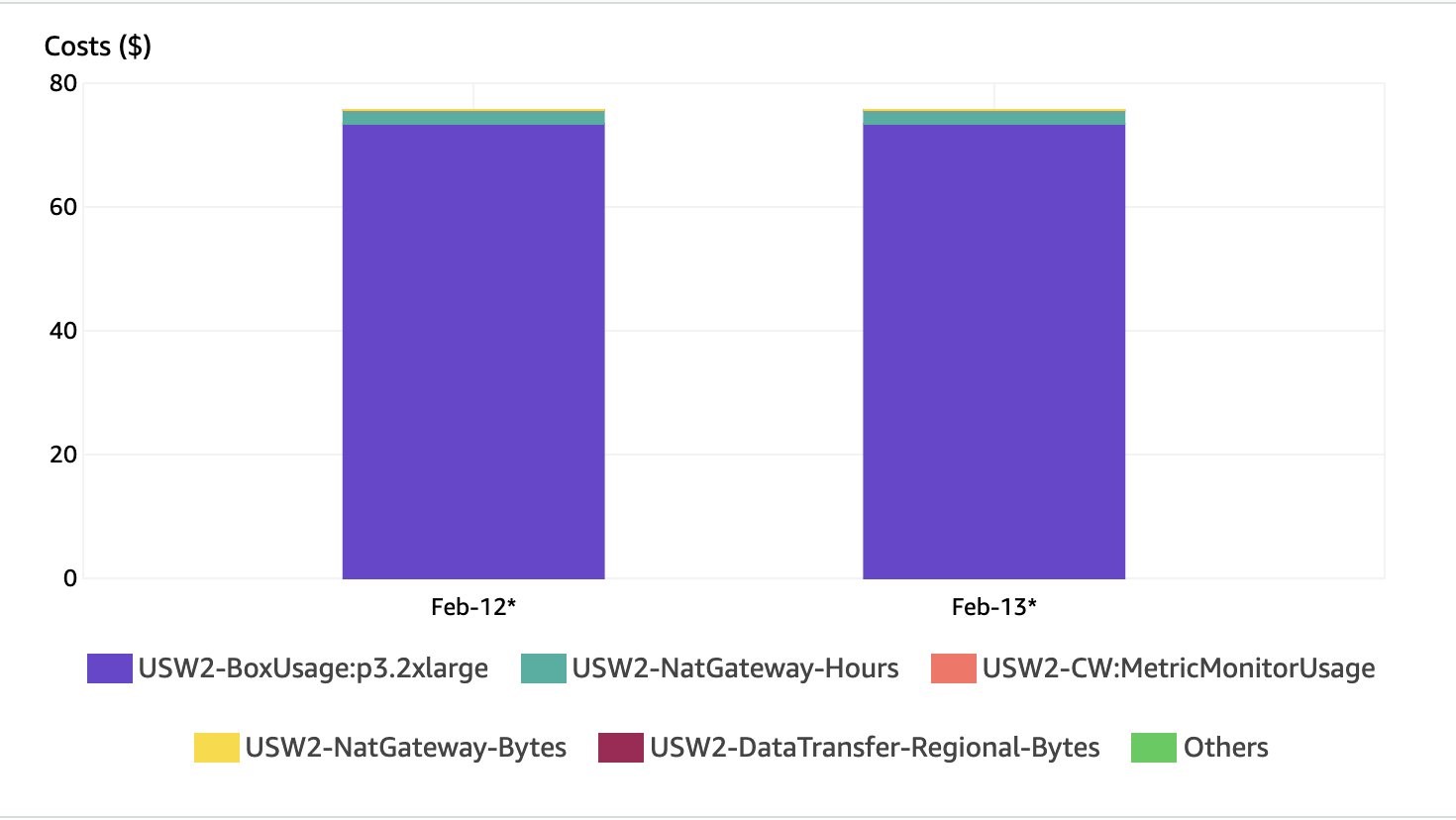
Running this solution with 1 instance costs approximately $75 a day in us-west-2.
Conclusion
This post described how to use NVIDIA MegaMolBART and CuChem on Amazon ECS containers to generate novel molecules for drug discovery. Amazon ECS and AWS CDK simplify the creation of the underlying infrastructure to execute containerized workloads at scale and enable repeatable deployments. AWS CDK supports multiple programming languages such as TypeScript, JavaScript, C#, Java, and Python, which many fellow scientists are already familiar with. We chose Python as the preferred language because of its popularity in data science. Combining the power and simplicity of Amazon ECS with containerized workloads, we aim to help AWS customers speed up research and lower the barriers to entry for development efforts for drug discovery. For deploying this service to your own AWS account, visit the AWS Quick Start page.
